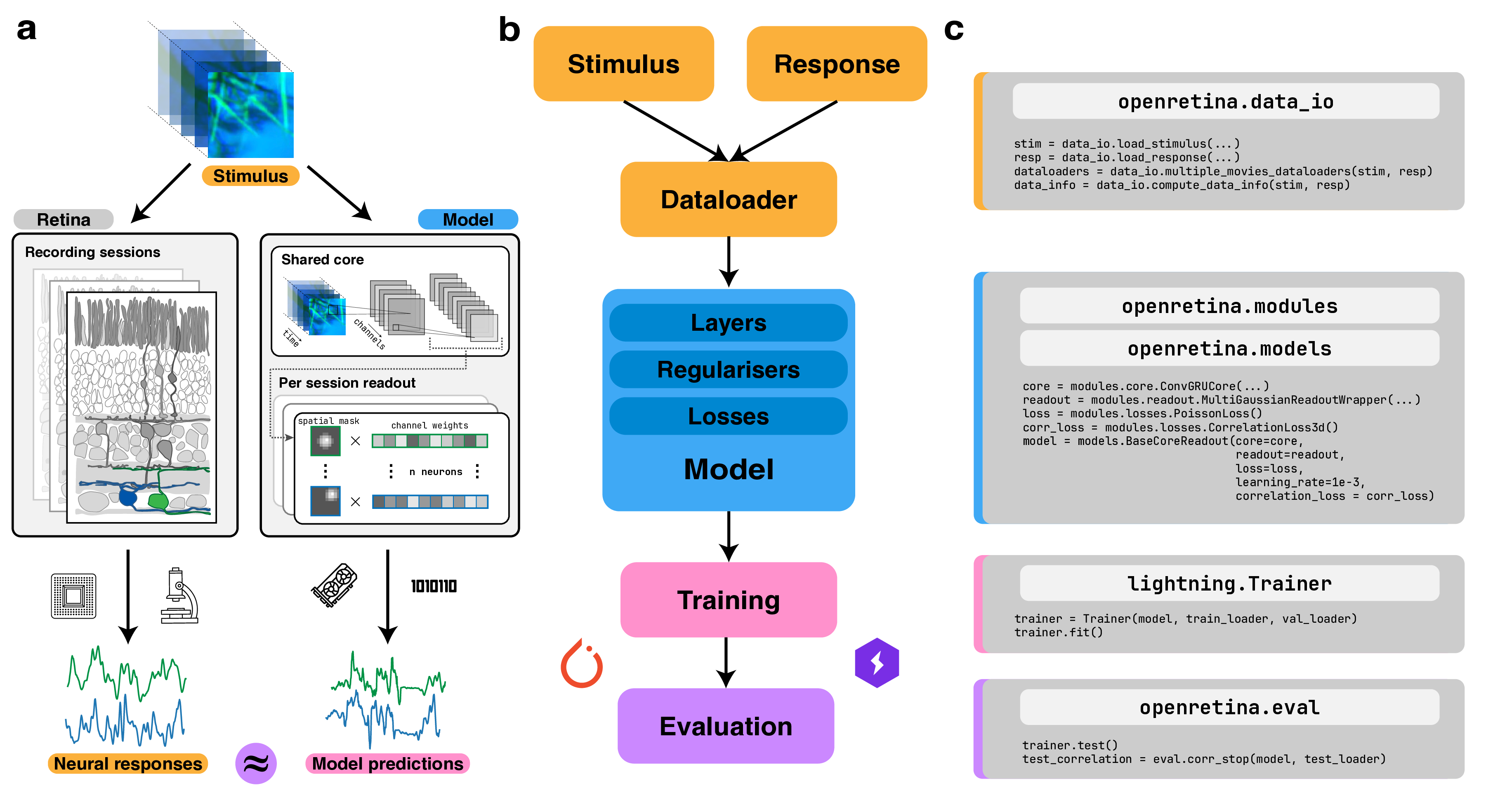
Training Overview
openretina builds on top of PyTorch, PyTorch Lightning, and Hydra to provide a flexible training stack. Lightning streamlines logging, checkpointing, and distributed execution, while Hydra keeps configuration files modular and composable. After a short learning curve, this combination lets you reproduce published models or explore new architectures with minimal boilerplate.
If you are preparing data for training, start with the data loading guide. When you are ready to deploy a specific model, the core + readout walkthrough and the training tutorial illustrate full experiments, including configuration files and command-line entry points.
What the Training Infrastructure Provides
- Configuration management: reusable Hydra configs under
configs/, including presets for datasets, models, and optimisation hyper-parameters. - Experiment tracking: automatic logging of losses, metrics, and checkpoints via PyTorch Lightning, with optional integrations for TensorBoard and WandB.
- Modular model components: interchangeable cores, readouts, regularisers, and losses from
openretina.modules, orchestrated throughopenretina.models. - Evaluation utilities: trained model evaluatino via
openretina.eval.
Typical Training Workflow
- Select data: Load stimuli and responses into dictionaries of train/validation/test splits as described in the Data I/O documentation.
- Choose configuration: Pick a base config (e.g.
configs/hoefling_2024_core_readout_low_res.yaml) and customise modules, hyper-parameters, or logging targets. - Launch training: Run
openretina train ...or the equivalent Python script. Lightning handles checkpoints, gradient accumulation, and mixed precision when enabled. - Monitor progress: Inspect metrics in TensorBoard or your preferred logger. Checkpoints are saved under the run directory specified by Hydra.
- Evaluate results: Use evaluation scripts or notebooks—--see the in-silico experiments overview for ideas—--to benchmark against ground truth or probe model behaviour.
The notebooks in notebooks/ mirror these steps with executable examples, including reproductions of the core + readout model on both calcium imaging and spiking datasets.